Statistics
Networks
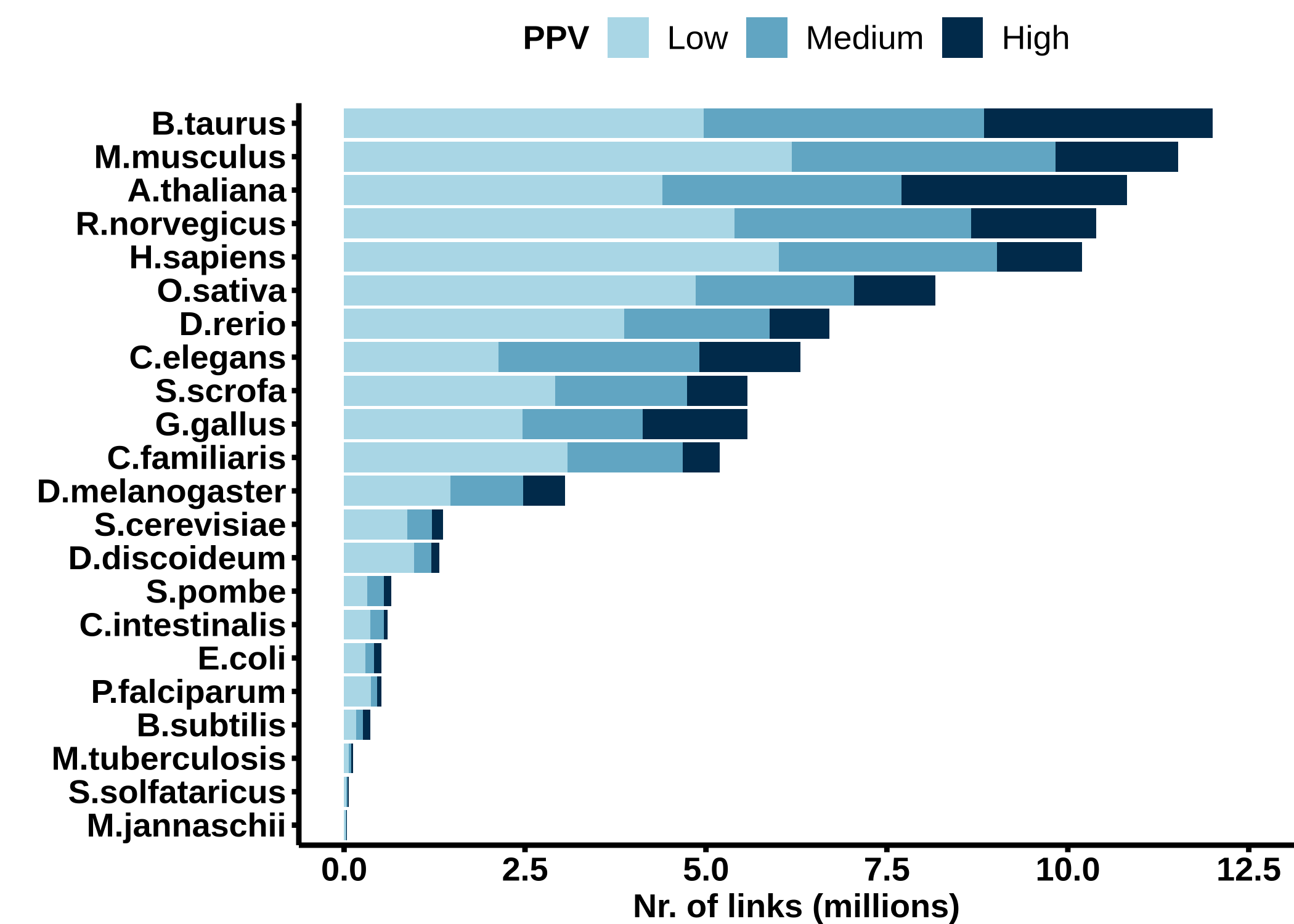
Sizes of the summary networks with the strongest links from each class for different ranges of final Bayesian scores (FBS). The fist interval corresponds roughly to a link confidence score (PPV) interval of 0.85 to 0.9 (Low), the second interval to 0.9 to 0.95 (Medium), and the last interval to 0.95 to 1 (High).
Coverage
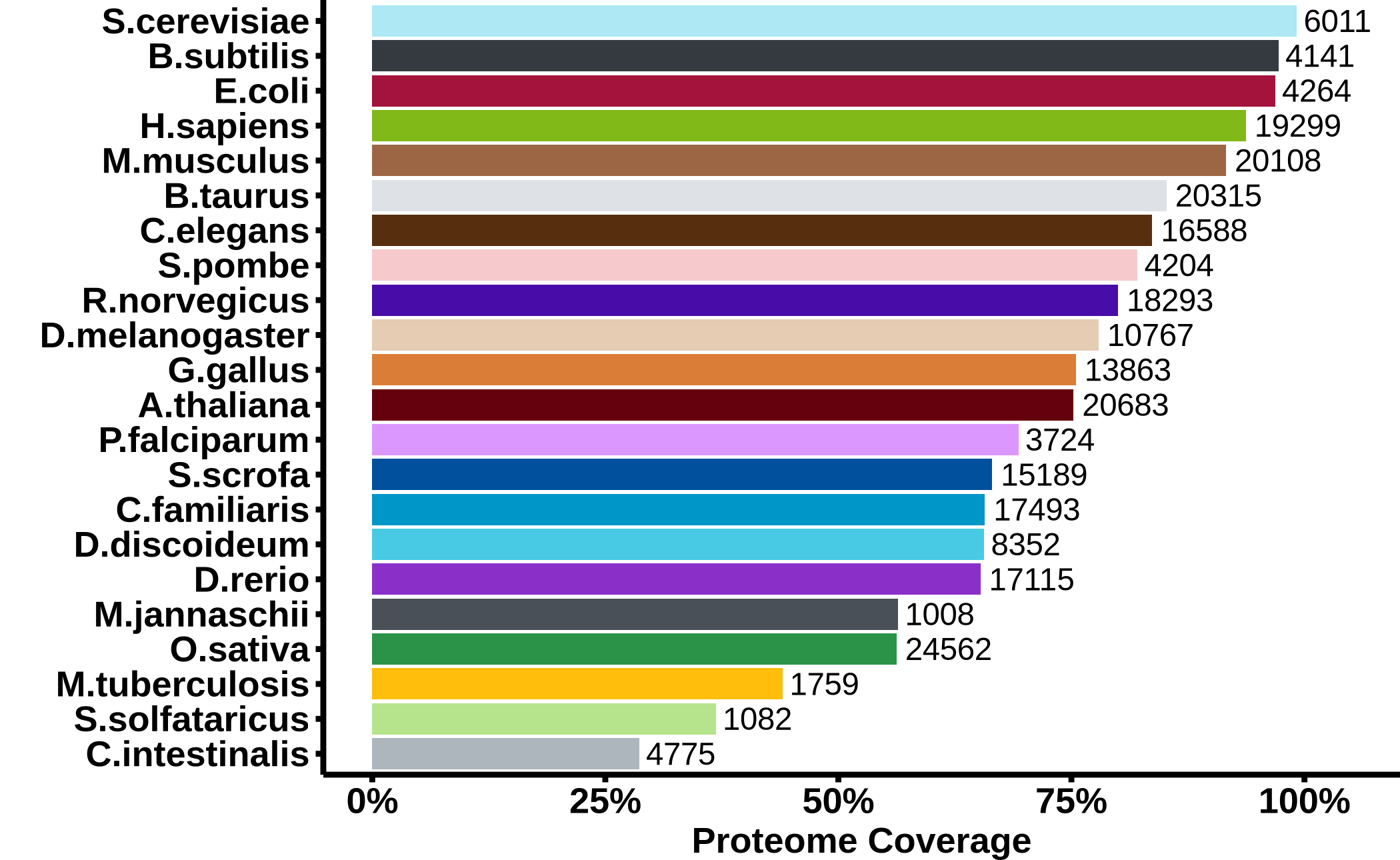
Number of genes (nodes) in the summary networks of the different species.
Gold Standards
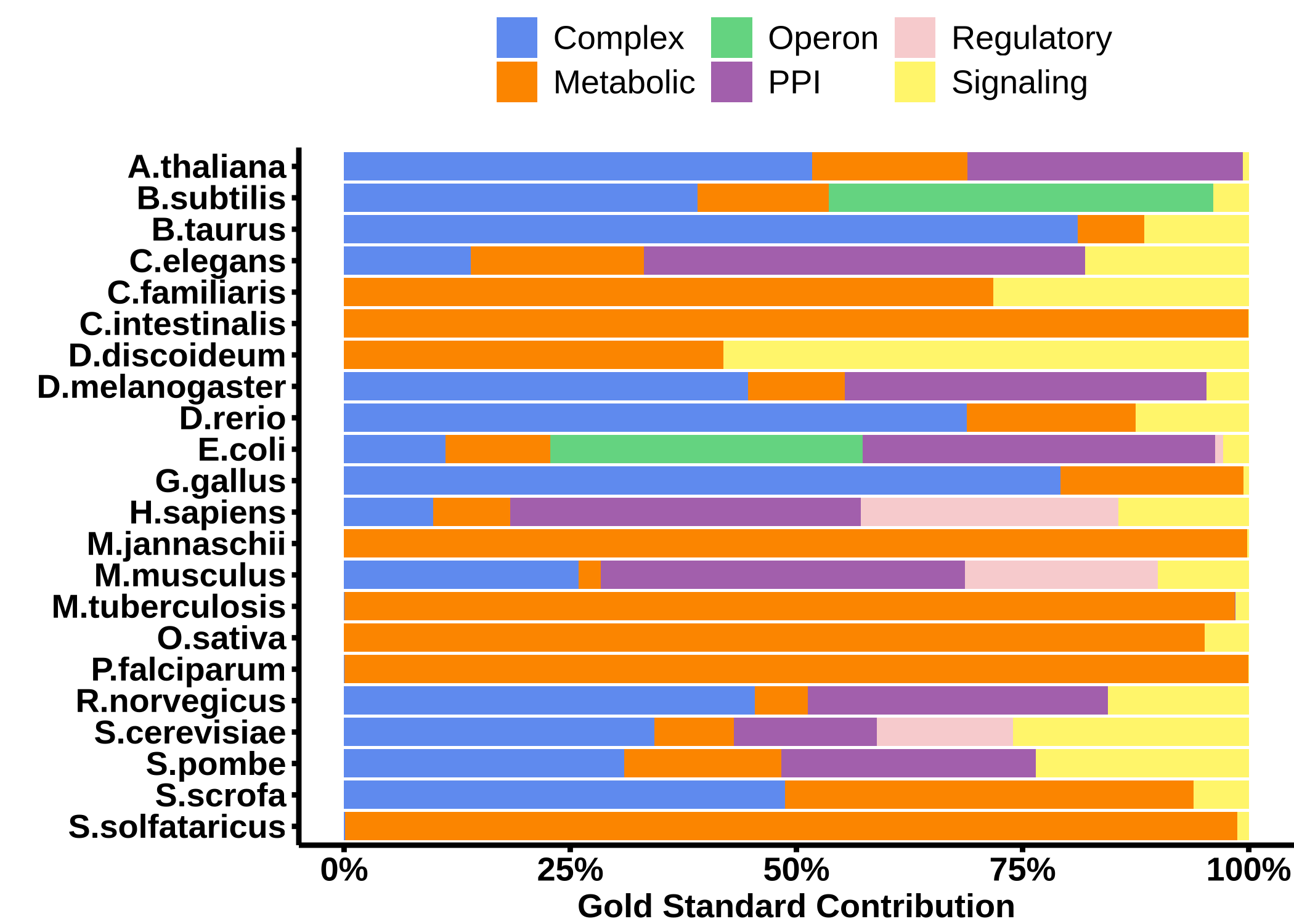
Contribution of Gold Standard to the networks of each FunCoup species. The number of links is normalized so it sums to 1 per species.
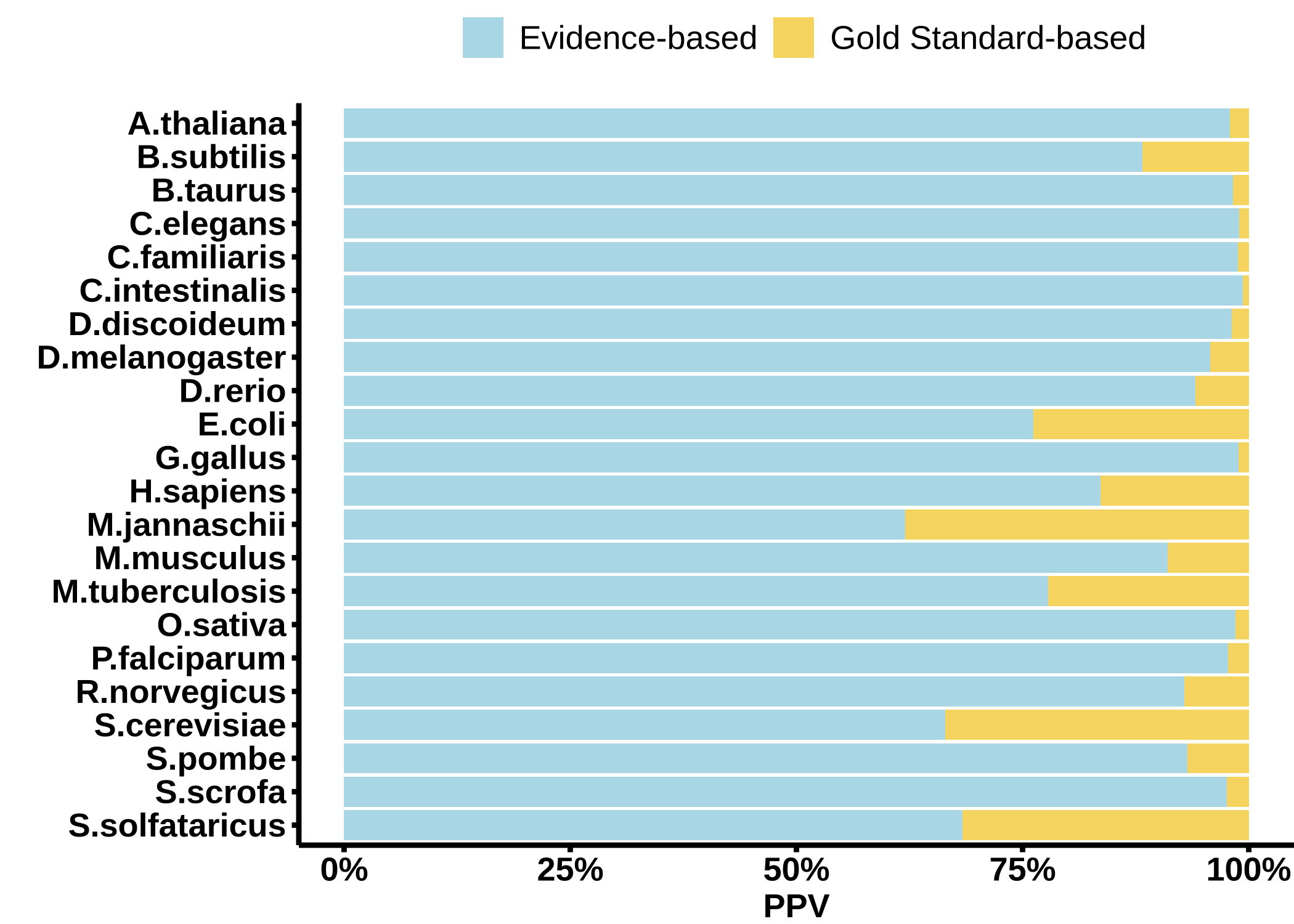
The percentage of links with Gold Standard- and Evidence-based confidence (PPV).
| Species | Type | Data | |
|---|---|---|---|
| Arabidopsis thaliana | Complex | 2022-08 | data source |
| Arabidopsis thaliana | Metabolic | 2022-02 | data source |
| Arabidopsis thaliana | PPI | 2022-08 | data source |
| Arabidopsis thaliana | Signaling | 2022-02 | data source |
| Bacillus subtilis (strain 168) | Complex | 2022-08 | data source |
| Bacillus subtilis (strain 168) | Metabolic | 2022-02 | data source |
| Bacillus subtilis (strain 168) | Operon | 2022-02 | data source |
| Bacillus subtilis (strain 168) | PPI | 2022-08 | data source |
| Bacillus subtilis (strain 168) | Signaling | 2022-02 | data source |
| Bos taurus | Complex | 2022-08 | data source |
| Bos taurus | Metabolic | 2022-02 | data source |
| Bos taurus | PPI | 2022-08 | data source |
| Bos taurus | Signaling | 2022-02 | data source |
| Caenorhabditis elegans | Complex | 2022-08 | data source |
| Caenorhabditis elegans | Metabolic | 2022-02 | data source |
| Caenorhabditis elegans | Operon | 2022-02 | data source |
| Caenorhabditis elegans | PPI | 2022-08 | data source |
| Caenorhabditis elegans | Signaling | 2022-02 | data source |
| Canis lupus familiaris | Complex | 2022-08 | data source |
| Canis lupus familiaris | Metabolic | 2022-02 | data source |
| Canis lupus familiaris | PPI | 2022-08 | data source |
| Canis lupus familiaris | Signaling | 2022-02 | data source |
| Ciona intestinalis | Complex | 2022-08 | data source |
| Ciona intestinalis | Metabolic | 2022-02 | data source |
| Ciona intestinalis | PPI | 2022-08 | data source |
| Ciona intestinalis | Signaling | 2022-02 | data source |
| Danio rerio | Complex | 2022-08 | data source |
| Danio rerio | Metabolic | 2022-02 | data source |
| Danio rerio | PPI | 2022-08 | data source |
| Danio rerio | Signaling | 2022-02 | data source |
| Dictyostelium discoideum | Complex | 2022-08 | data source |
| Dictyostelium discoideum | Metabolic | 2022-02 | data source |
| Dictyostelium discoideum | PPI | 2022-08 | data source |
| Dictyostelium discoideum | Signaling | 2022-02 | data source |
| Drosophila melanogaster | Complex | 2022-08 | data source |
| Drosophila melanogaster | Metabolic | 2022-02 | data source |
| Drosophila melanogaster | PPI | 2022-08 | data source |
| Drosophila melanogaster | Signaling | 2022-02 | data source |
| Escherichia coli (strain K12) | Complex | 2022-08 | data source |
| Escherichia coli (strain K12) | Metabolic | 2022-02 | data source |
| Escherichia coli (strain K12) | Operon | 2022-02 | data source |
| Escherichia coli (strain K12) | PPI | 2022-08 | data source |
| Escherichia coli (strain K12) | Regulatory | 2022-12 | data source |
| Escherichia coli (strain K12) | Signaling | 2022-02 | data source |
| Gallus gallus | Complex | 2022-08 | data source |
| Gallus gallus | Metabolic | 2022-02 | data source |
| Gallus gallus | PPI | 2022-08 | data source |
| Gallus gallus | Signaling | 2022-02 | data source |
| Homo sapiens | Complex | 2022-08 | data source |
| Homo sapiens | Metabolic | 2022-02 | data source |
| Homo sapiens | PPI | 2022-08 | data source |
| Homo sapiens | Regulatory | 2019-04,2018-04 | data source |
| Homo sapiens | Signaling | 2022-02 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | Complex | 2022-08 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | Metabolic | 2022-02 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | PPI | 2022-08 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | Signaling | 2022-02 | data source |
| Mus musculus | Complex | 2022-08 | data source |
| Mus musculus | Metabolic | 2022-02 | data source |
| Mus musculus | PPI | 2022-08 | data source |
| Mus musculus | Regulatory | 2019-04,2018-04 | data source |
| Mus musculus | Signaling | 2022-02 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | Complex | 2022-08 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | Metabolic | 2022-02 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | PPI | 2022-08 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | Signaling | 2022-02 | data source |
| Oryza sativa (subsp. japonica) | Complex | 2022-08 | data source |
| Oryza sativa (subsp. japonica) | Metabolic | 2022-02 | data source |
| Oryza sativa (subsp. japonica) | PPI | 2022-08 | data source |
| Oryza sativa (subsp. japonica) | Signaling | 2022-02 | data source |
| Plasmodium falciparum | Complex | 2022-08 | data source |
| Plasmodium falciparum | Metabolic | 2022-02 | data source |
| Plasmodium falciparum | PPI | 2022-08 | data source |
| Plasmodium falciparum | Signaling | 2022-02 | data source |
| Rattus norvegicus | Complex | 2022-08 | data source |
| Rattus norvegicus | Metabolic | 2022-02 | data source |
| Rattus norvegicus | PPI | 2022-08 | data source |
| Rattus norvegicus | Signaling | 2022-02 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | Complex | 2022-08 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | Metabolic | 2022-02 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | PPI | 2022-08 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | Signaling | 2022-02 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | Complex | 2022-08 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | Metabolic | 2022-02 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | PPI | 2022-08 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | Regulatory | 2022 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | Signaling | 2022-02 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | Complex | 2022-08 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | Metabolic | 2022-02 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | PPI | 2022-08 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | Signaling | 2022-02 | data source |
| Sus scrofa | Complex | 2022-08 | data source |
| Sus scrofa | Metabolic | 2022-02 | data source |
| Sus scrofa | PPI | 2022-08 | data source |
| Sus scrofa | Signaling | 2022-02 | data source |
Evidences
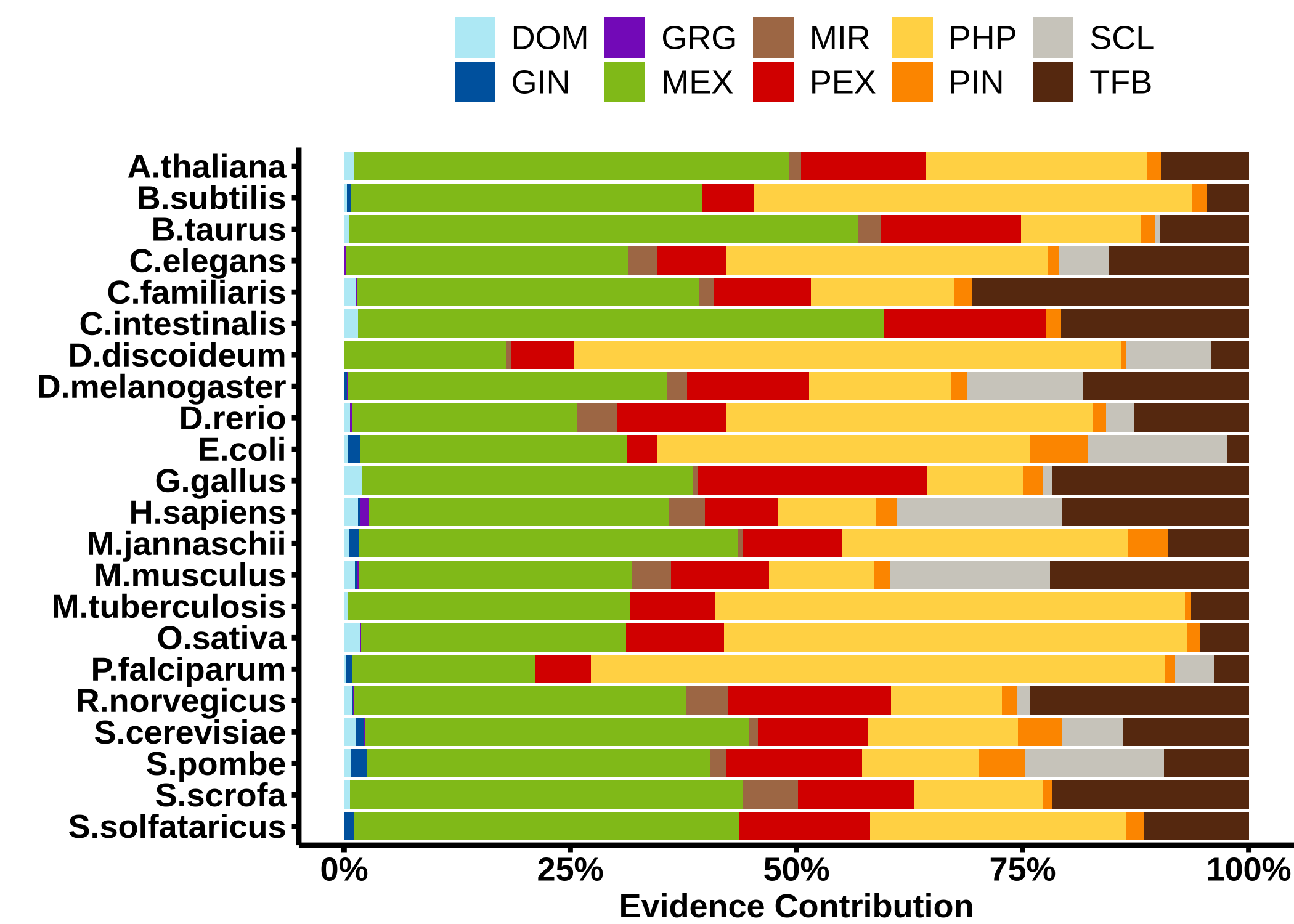
Relative impact of the different evidence types as a fraction of the total FBS for the different networks. The different types are: domain interactions (DOM), genetic interaction profile similarity (GIN), gene regulation (GRG), mRNA co-expression (MEX), co-miRNA regulation by shared miRNA targeting (MIR), protein co-expression (PEX), phylogenetic profile similarity (PHP), protein-protein interaction (PPI), sub-cellular co-localization (SCL), and shared transcription factor binding (TFB).
| Species | Type | Data | |
|---|---|---|---|
| Arabidopsis thaliana | DOM | UniDomInt_v1.0 | data source |
| Arabidopsis thaliana | MEX | E-GEOD-30720 | data source |
| Arabidopsis thaliana | MEX | E-GEOD-53197 | data source |
| Arabidopsis thaliana | MEX | E-MTAB-7978 | data source |
| Arabidopsis thaliana | MEX | E-MTAB-7933 | data source |
| Arabidopsis thaliana | MEX | E-CURD-1 | data source |
| Arabidopsis thaliana | PEX | PaxDb_v5.0 | data source |
| Arabidopsis thaliana | PHP | InParanoiDB_v9.0 | data source |
| Arabidopsis thaliana | PIN | iRefIndex_v2022-08 | data source |
| Arabidopsis thaliana | SCL | GeneOntology_v2023-03 | data source |
| Bacillus subtilis (strain 168) | DOM | UniDomInt_v1.0 | data source |
| Bacillus subtilis (strain 168) | MEX | GSE69575 | data source |
| Bacillus subtilis (strain 168) | MEX | GSE27219 | data source |
| Bacillus subtilis (strain 168) | MEX | GSE19831 | data source |
| Bacillus subtilis (strain 168) | MEX | GSE44125 | data source |
| Bacillus subtilis (strain 168) | PHP | InParanoiDB_v9.0 | data source |
| Bacillus subtilis (strain 168) | PIN | iRefIndex_v2022-08 | data source |
| Bacillus subtilis (strain 168) | SCL | GeneOntology_v2023-03 | data source |
| Bos taurus | DOM | UniDomInt_v1.0 | data source |
| Bos taurus | MEX | E-MTAB-2596 | data source |
| Bos taurus | MEX | E-MTAB-2798 | data source |
| Bos taurus | MEX | E-MTAB-7887 | data source |
| Bos taurus | PHP | InParanoiDB_v9.0 | data source |
| Bos taurus | PIN | iRefIndex_v2022-08 | data source |
| Bos taurus | SCL | GeneOntology_v2023-03 | data source |
| Caenorhabditis elegans | DOM | UniDomInt_v1.0 | data source |
| Caenorhabditis elegans | GIN | BioGRID_v4.4.219 | data source |
| Caenorhabditis elegans | GRG | ENCODE_v131.0 | data source |
| Caenorhabditis elegans | MEX | E-MTAB-2812 | data source |
| Caenorhabditis elegans | MIR | microRNA.org_v2010 | data source |
| Caenorhabditis elegans | PHP | InParanoiDB_v9.0 | data source |
| Caenorhabditis elegans | PIN | iRefIndex_v2022-08 | data source |
| Caenorhabditis elegans | SCL | GeneOntology_v2023-03 | data source |
| Caenorhabditis elegans | TFB | TFLink_v1.0 | data source |
| Canis lupus familiaris | DOM | UniDomInt_v1.0 | data source |
| Canis lupus familiaris | MEX | GSE20113 | data source |
| Canis lupus familiaris | MEX | GSE39005 | data source |
| Canis lupus familiaris | MEX | GSE23760 | data source |
| Canis lupus familiaris | PHP | InParanoiDB_v9.0 | data source |
| Canis lupus familiaris | PIN | iRefIndex_v2022-08 | data source |
| Canis lupus familiaris | SCL | GeneOntology_v2023-03 | data source |
| Ciona intestinalis | DOM | UniDomInt_v1.0 | data source |
| Ciona intestinalis | PHP | InParanoiDB_v9.0 | data source |
| Ciona intestinalis | PIN | iRefIndex_v2022-08 | data source |
| Danio rerio | DOM | UniDomInt_v1.0 | data source |
| Danio rerio | MEX | E-ERAD-475 | data source |
| Danio rerio | PEX | PaxDb_v5.0 | data source |
| Danio rerio | PHP | InParanoiDB_v9.0 | data source |
| Danio rerio | PIN | iRefIndex_v2022-08 | data source |
| Danio rerio | SCL | GeneOntology_v2023-03 | data source |
| Danio rerio | TFB | TFLink_v1.0 | data source |
| Dictyostelium discoideum | DOM | UniDomInt_v1.0 | data source |
| Dictyostelium discoideum | MEX | GSE57212 | data source |
| Dictyostelium discoideum | MEX | GSE71036 | data source |
| Dictyostelium discoideum | MEX | GSE8287 | data source |
| Dictyostelium discoideum | PHP | InParanoiDB_v9.0 | data source |
| Dictyostelium discoideum | PIN | iRefIndex_v2022-08 | data source |
| Dictyostelium discoideum | SCL | GeneOntology_v2023-03 | data source |
| Drosophila melanogaster | DOM | UniDomInt_v1.0 | data source |
| Drosophila melanogaster | GRG | ENCODE_v131.0 | data source |
| Drosophila melanogaster | MEX | E-GEOD-20348 | data source |
| Drosophila melanogaster | MEX | GSE38106 | data source |
| Drosophila melanogaster | MEX | GSE11316 | data source |
| Drosophila melanogaster | MEX | GSE23320 | data source |
| Drosophila melanogaster | MEX | E-GEOD-18068 | data source |
| Drosophila melanogaster | MIR | microRNA.org_v2010 | data source |
| Drosophila melanogaster | PHP | InParanoiDB_v9.0 | data source |
| Drosophila melanogaster | PIN | iRefIndex_v2022-08 | data source |
| Drosophila melanogaster | SCL | GeneOntology_v2023-03 | data source |
| Drosophila melanogaster | TFB | TFLink_v1.0 | data source |
| Escherichia coli (strain K12) | DOM | UniDomInt_v1.0 | data source |
| Escherichia coli (strain K12) | GIN | BioGRID_v4.4.219 | data source |
| Escherichia coli (strain K12) | MEX | GSE40811 | data source |
| Escherichia coli (strain K12) | MEX | GSE20305 | data source |
| Escherichia coli (strain K12) | MEX | GSE7265 | data source |
| Escherichia coli (strain K12) | PHP | InParanoiDB_v9.0 | data source |
| Escherichia coli (strain K12) | PIN | iRefIndex_v2022-08 | data source |
| Escherichia coli (strain K12) | SCL | GeneOntology_v2023-03 | data source |
| Gallus gallus | DOM | UniDomInt_v1.0 | data source |
| Gallus gallus | MEX | E-MTAB-2797 | data source |
| Gallus gallus | MEX | E-MTAB-7663 | data source |
| Gallus gallus | MEX | E-MTAB-6769 | data source |
| Gallus gallus | MEX | GSE10538 | data source |
| Gallus gallus | PEX | PaxDb_v5.0 | data source |
| Gallus gallus | PHP | InParanoiDB_v9.0 | data source |
| Gallus gallus | PIN | iRefIndex_v2022-08 | data source |
| Gallus gallus | SCL | GeneOntology_v2023-03 | data source |
| Homo sapiens | DOM | UniDomInt_v1.0 | data source |
| Homo sapiens | GIN | BioGRID_v4.4.219 | data source |
| Homo sapiens | GRG | ENCODE_v131.0 | data source |
| Homo sapiens | MEX | E-MTAB-1733 | data source |
| Homo sapiens | MEX | E-MTAB-2836 | data source |
| Homo sapiens | MEX | E-MTAB-6814 | data source |
| Homo sapiens | MEX | E-ENAD-34 | data source |
| Homo sapiens | MEX | E-MTAB-5214 | data source |
| Homo sapiens | MIR | microRNA.org_v2010 | data source |
| Homo sapiens | PEX | PaxDb_v5.0 | data source |
| Homo sapiens | PHP | InParanoiDB_v9.0 | data source |
| Homo sapiens | PIN | iRefIndex_v2022-08 | data source |
| Homo sapiens | SCL | GeneOntology_v2023-03 | data source |
| Homo sapiens | TFB | TFLink_v1.0 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | DOM | UniDomInt_v1.0 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | MEX | GSE4620 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | PHP | InParanoiDB_v9.0 | data source |
| Methanocaldococcus jannaschii (strain ATCC 43067 / DSM 2661 / JAL-1 / JCM 10045 / NBRC 100440) | PIN | iRefIndex_v2022-08 | data source |
| Mus musculus | DOM | UniDomInt_v1.0 | data source |
| Mus musculus | GRG | ENCODE_v131.0 | data source |
| Mus musculus | MEX | E-MTAB-3718 | data source |
| Mus musculus | MEX | E-MTAB-2801 | data source |
| Mus musculus | MEX | E-GEOD-70484 | data source |
| Mus musculus | MEX | E-MTAB-6798 | data source |
| Mus musculus | MIR | microRNA.org_v2010 | data source |
| Mus musculus | PEX | PaxDb_v5.0 | data source |
| Mus musculus | PHP | InParanoiDB_v9.0 | data source |
| Mus musculus | PIN | iRefIndex_v2022-08 | data source |
| Mus musculus | SCL | GeneOntology_v2023-03 | data source |
| Mus musculus | TFB | TFLink_v1.0 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | DOM | UniDomInt_v1.0 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | PHP | InParanoiDB_v9.0 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | PIN | iRefIndex_v2022-08 | data source |
| Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) | SCL | GeneOntology_v2023-03 | data source |
| Oryza sativa (subsp. japonica) | DOM | UniDomInt_v1.0 | data source |
| Oryza sativa (subsp. japonica) | PHP | InParanoiDB_v9.0 | data source |
| Oryza sativa (subsp. japonica) | PIN | iRefIndex_v2022-08 | data source |
| Oryza sativa (subsp. japonica) | SCL | GeneOntology_v2023-03 | data source |
| Plasmodium falciparum | DOM | UniDomInt_v1.0 | data source |
| Plasmodium falciparum | MEX | GSE59097 | data source |
| Plasmodium falciparum | MEX | GSE59099 | data source |
| Plasmodium falciparum | PHP | InParanoiDB_v9.0 | data source |
| Plasmodium falciparum | PIN | iRefIndex_v2022-08 | data source |
| Plasmodium falciparum | SCL | GeneOntology_v2023-03 | data source |
| Rattus norvegicus | DOM | UniDomInt_v1.0 | data source |
| Rattus norvegicus | MEX | E-MTAB-2800 | data source |
| Rattus norvegicus | MEX | GSE63362 | data source |
| Rattus norvegicus | MEX | E-MTAB-6811 | data source |
| Rattus norvegicus | MEX | E-GEOD-53960 | data source |
| Rattus norvegicus | MIR | microRNA.org_v2010 | data source |
| Rattus norvegicus | PEX | PaxDb_v5.0 | data source |
| Rattus norvegicus | PHP | InParanoiDB_v9.0 | data source |
| Rattus norvegicus | PIN | iRefIndex_v2022-08 | data source |
| Rattus norvegicus | SCL | GeneOntology_v2023-03 | data source |
| Rattus norvegicus | TFB | TFLink_v1.0 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | DOM | UniDomInt_v1.0 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | MEX | GSE12235 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | PHP | InParanoiDB_v9.0 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | PIN | iRefIndex_v2022-08 | data source |
| Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2) | SCL | GeneOntology_v2023-03 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | DOM | UniDomInt_v1.0 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | GIN | BioGRID_v4.4.219 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | MEX | GSE33099 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | MEX | GSE33097 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | MEX | GSE26169 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | MEX | GSE8561 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | PHP | InParanoiDB_v9.0 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | PIN | iRefIndex_v2022-08 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | SCL | GeneOntology_v2023-03 | data source |
| Saccharomyces cerevisiae (strain ATCC 204508 / S288c) | TFB | TFLink_v1.0 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | DOM | UniDomInt_v1.0 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | GIN | BioGRID_v4.4.219 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | MEX | GSE4284 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | PHP | InParanoiDB_v9.0 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | PIN | iRefIndex_v2022-08 | data source |
| Schizosaccharomyces pombe (strain 972 / ATCC 24843) | SCL | GeneOntology_v2023-03 | data source |
| Sus scrofa | DOM | UniDomInt_v1.0 | data source |
| Sus scrofa | MEX | E-MTAB-5895 | data source |
| Sus scrofa | MEX | GSE97374 | data source |
| Sus scrofa | MEX | GSE83932 | data source |
| Sus scrofa | PHP | InParanoiDB_v9.0 | data source |
| Sus scrofa | PIN | iRefIndex_v2022-08 | data source |
| Sus scrofa | SCL | GeneOntology_v2023-03 | data source |
Species
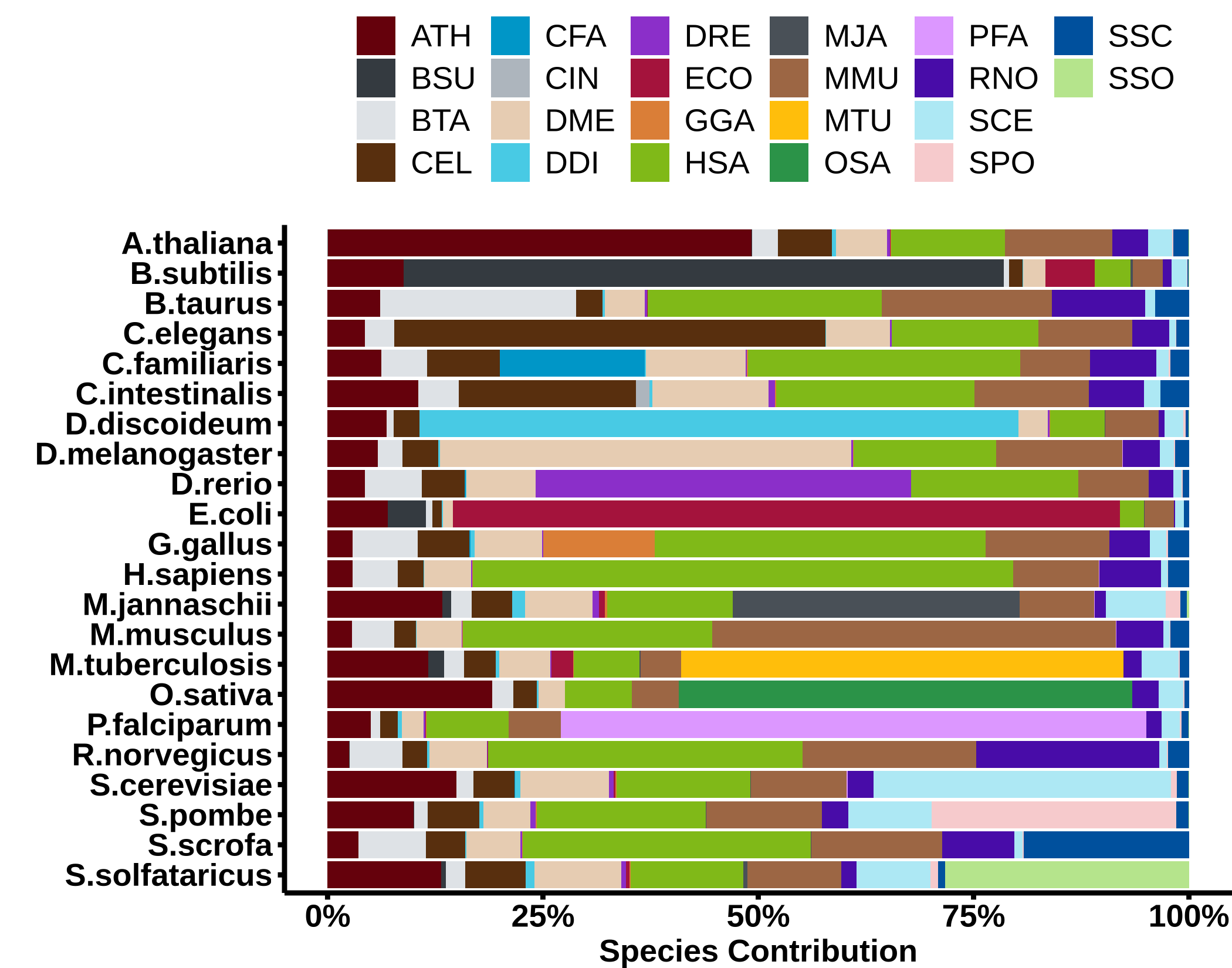
Relative impact of the evidence from the different species as a fraction of the total FBS. For most species more than 50% of the evidence comes from other species.